PUBLICATIONS
Please refer to the Google Scholar page for an updated list of publications.
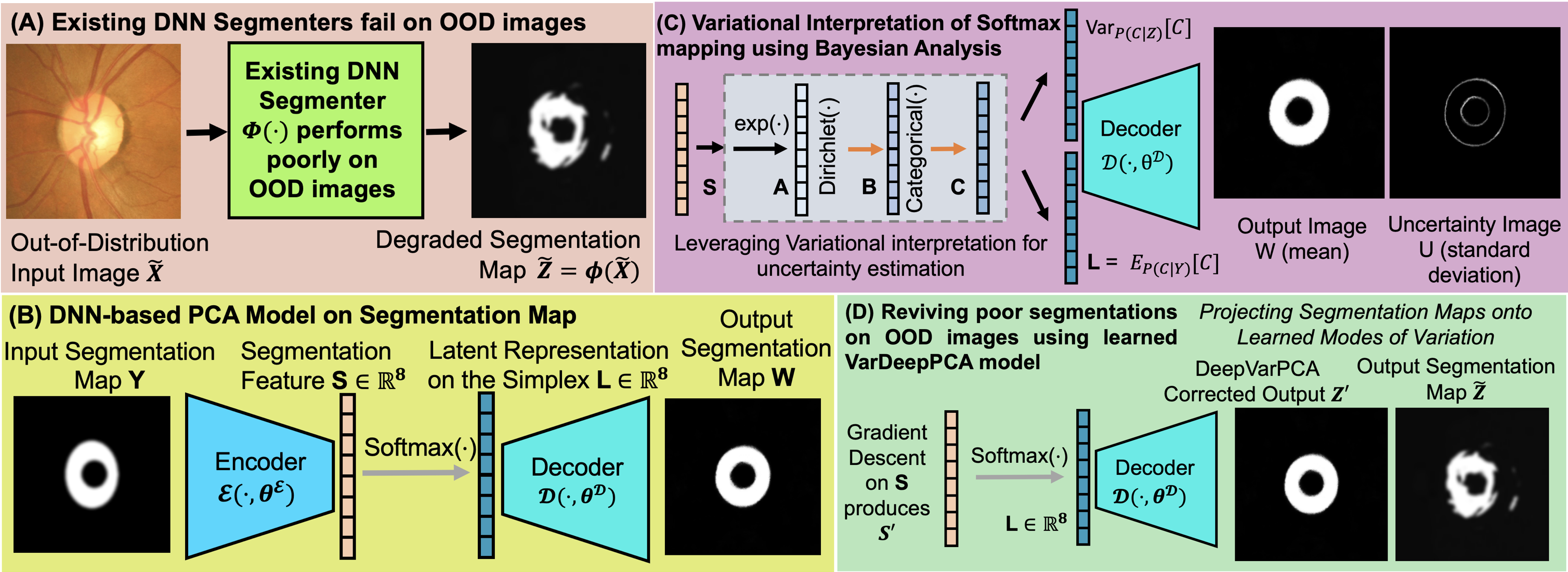
Reviving Poor Object Segmentations in OOD Medical Images using Variational-Deep-PCA Modeling on Segmentation Maps with Sampling-Free Learning
Winter Conference on Applications of Computer Vision (WACV) 2024
Abstract: For object segmentation in medical images deep neural networks (DNNs) typically perform poorly on out-of-distribution (OOD) images stemming from the large variability in image-acquisition equipment and protocols across sites. However compared to such variability in the acquired medical images we observe that the variability in the underlying object-segmentation maps is far lower. Thus we propose a novel DNN framework to model this variability in segmentation maps and leverage it to revive poor segmentations produced by existing DNNs on OOD images. Our DNN framework (i) learns the principal modes of variation in a class of segmentation maps (ii) models each segmentation map using a low-dimensional mixture-of-modes latent representation on a simplex (iii) enables sampling-free variational learning and uncertainty estimation and (iv) trains using small in-distribution image sets. When OOD-image segmentations are extremely poor we propose a novel human-in-the-loop method needing minuscule human intervention. Results using 6 publicly-available datasets and 8 existing DNN segmenters show the benefits of our framework in OOD-image object segmentation.
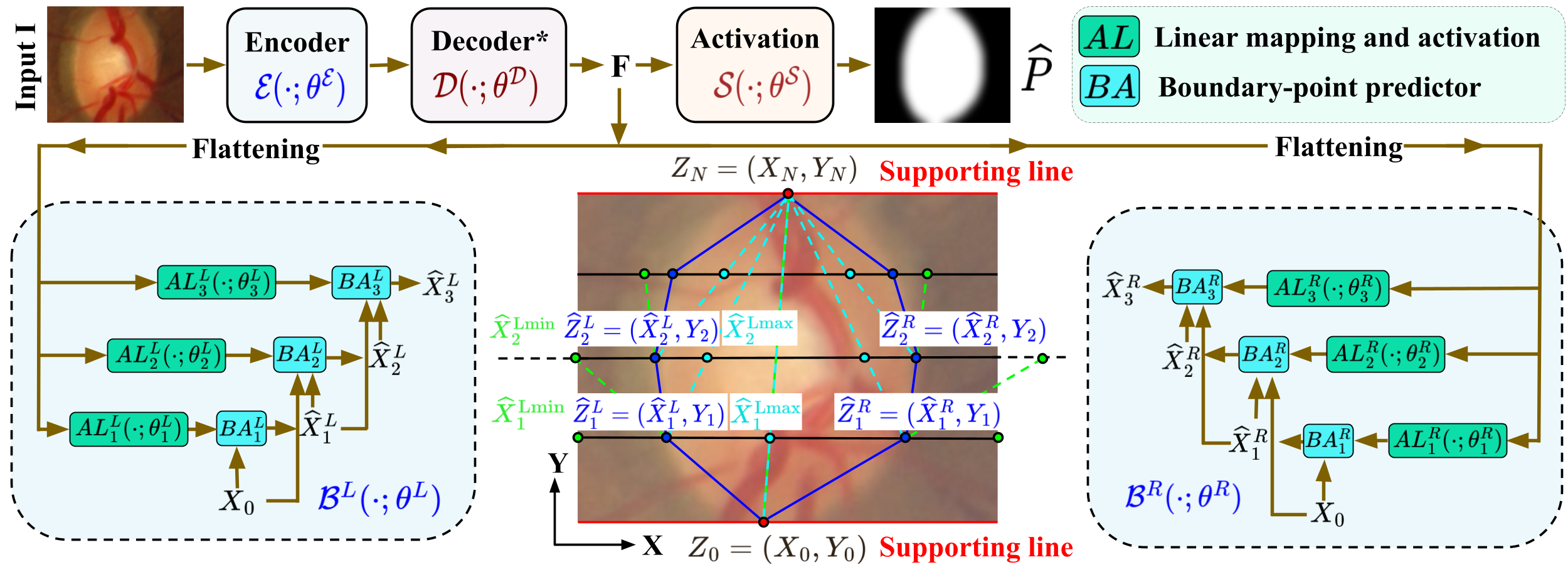
Convex Segments for Convex Objects using DNN Boundary Tracing and Graduated Optimization
International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2024.
Abstract: Image segmentation often involves objects of interest that are biologically known to be convex shaped. While typical deep-neural-networks (DNNs) for object segmentation ignore object properties relating to shape, the DNNs that employ shape information fail to enforce hard constraints on shape. We design a brand-new DNN framework that guarantees convexity of the output object-segment by leveraging fundamental geometrical insights into the boundaries of convex-shaped objects. Moreover, we design our framework to build on typical existing DNNs for per-pixel segmentation, while maintaining simplicity in loss-term formulation and maintaining frugality in model size and training time. Results using six publicly available datasets demonstrates that our DNN framework, with little overheads, provides significant benefits in the robust segmentation of convex objects in out-of-distribution images.
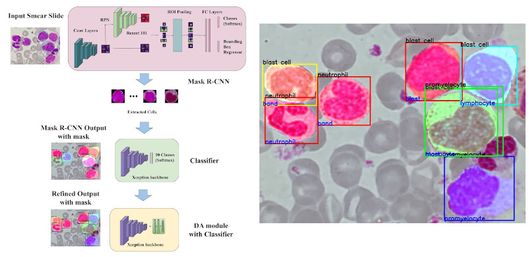
Advancing instance segmentation and WBC classification in peripheral blood smear through domain adaptation - A study on PBC and the novel RV-PBS datasets
Abstract: Automating blood cell counting and detection from smear slides holds significant potential for aiding doctors in disease diagnosis through blood tests. However, existing literature has not adequately addressed using whole slide data in this context. This study introduces the novel RV-PBS dataset, comprising ten distinct peripheral blood smear classes, each featuring multiple multi-class White Blood Cells per slide, specifically designed, for instance segmentation benchmarks. While conventional instance segmentation models like Mask R-CNN exhibit promising results in segmenting medical artifact instances, they face challenges in scenarios with limited samples and class imbalances within the dataset. This challenge prompted us to explore innovative techniques such as domain adaptation using a similar dataset to enhance the classification accuracy of Mask R-CNN, a novel approach in the domain of medical image analysis. Our study has successfully established a comprehensive pipeline capable of segmenting, detecting, and classifying blood samples from slides, striking an optimal balance between computational complexity and accurate classification of medical artifacts. This advancement enables precise cell counting and classification, facilitating doctors in refining their diagnostic analyses.
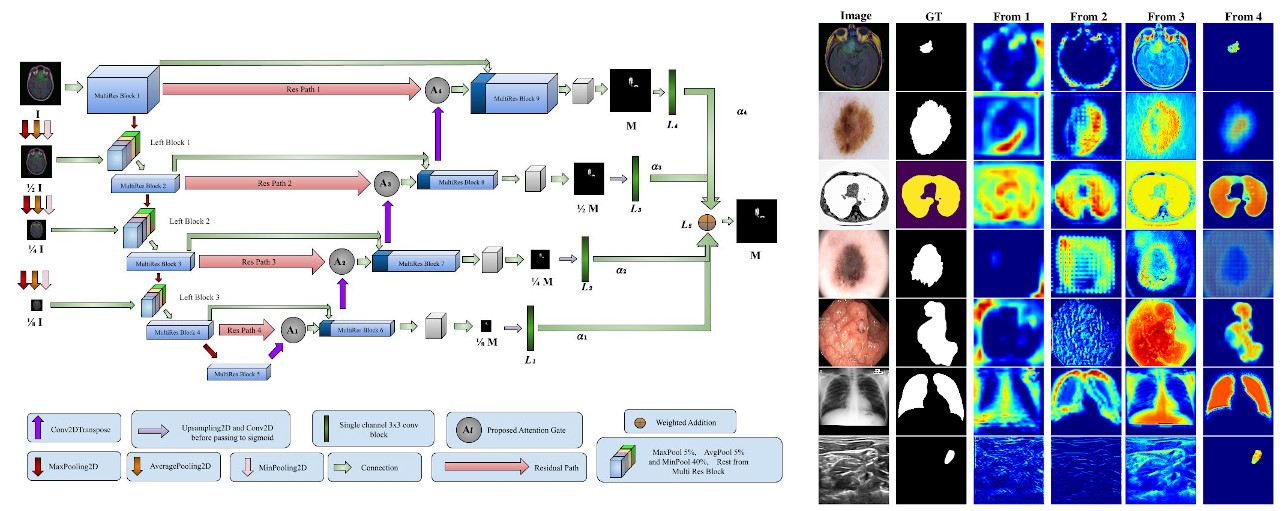
Improving Multi Scale Attention Networks - Bayesian Optimization for Segmenting medical images
The Imaging Science Journal. (2023)
Abstract: Current deep learning based image segmentation methods are notable for their use of large number of parameters and extensive computational resources in training. There is a persistent need for more efficient flexible systems without compromising on precision. This work proposes a novel model that combines the best of deep learning and probabilistic machine learning to segment a wide variety of medical image datasets with state-of-the-art accuracy and limited resources. The approach benefits from the introduction of new diverse attention modules that serve multiple purposes including capturing of relevant information at different scales. These proposed attention modules are generic and can potentially be used with other architectures to boost performance. In addition, Bayesian optimization is employed to tune multi-scale weight hyperparameters of the model. The architecture combined with one of the proposed novel attention modules and tuned hyperparameters achieves the best results in segmenting ISIC 2017, LUNGS, NERVE, Skin Lesion, and CHEST datasets. Finally, the explainability of the network is analyzed by visualizing the feature map learned from the attention modules.

Biomedical image analysis competitions - The state of current participation practice
arXiv. (2022)
Abstract: The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. This holds especially true for the field of biomedical image analysis, for which dozens of competitions are organized each year. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development for biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) and the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in the year 2021 (n = 80 competitions in total). Besides questions pertaining to general information on the team and the tackled tasks, the survey covered participants’ expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). Surprisingly, only 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. KSurprisingly, k-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Matthias Eisenmann, Annika Reinke, Vivienn Weru, Minu Dietlinde Tizabi, Fabian Isensee, Tim J. Adler, Patrick Godau, Veronika Cheplygina, Michal Kozubek, Sharib Ali, Anubha Gupta, Jan Kybic, Alison Noble, Carlos Ortiz de Solórzano, Samiksha Pachade, Caroline Petitjean, Daniel Sage, Donglai Wei, Elizabeth Wilden, Deepak Alapatt, Vincent Andrearczyk, Ujjwal Baid, Spyridon Bakas, Niranjan Balu, Sophia Bano, Vivek Singh Bawa, Jorge Bernal, Sebastian Bodenstedt, Alessandro Casella, Jinwook Choi, Olivier Commowick, Marie Daum, Adrien Depeursinge, Reuben Dorent, Jan Egger, Hannah Eichhorn, Sandy Engelhardt, Melanie Ganz, Gabriel Girard, Lasse Hansen, Mattias Heinrich, Nicholas Heller, Alessa Hering, Arnaud Huaulmé, Hyunjeong Kim, Bennett Landman, Hongwei Bran Li, Jianning Li, Jun Ma, Anne Martel, Carlos Martín-Isla, Bjoern Menze, Chinedu Innocent Nwoye, Valentin Oreiller, Nicolas Padoy, Sarthak Pati, Kelly Payette, Carole Sudre, Kimberlin van Wijnen, Armine Vardazaryan, Tom Vercauteren, Martin Wagner, Chuanbo Wang, Moi Hoon Yap, Zeyun Yu, Chun Yuan, Maximilian Zenk, Aneeq Zia, David Zimmerer, Rina Bao, Chanyeol Choi, Andrew Cohen, Oleh Dzyubachyk, Adrian Galdran, Tianyuan Gan, Tianqi Guo, Pradyumna Gupta, Mahmood Haithami, Edward Ho, Ikbeom Jang, Zhili Li, Zhengbo Luo, Filip Lux, Sokratis Makrogiannis, Dominik Müller, Young-tack Oh, Subeen Pang, Constantin Pape, Gorkem Polat, Charlotte Rosalie Reed, Kanghyun Ryu, Tim Scherr, Vajira Thambawita, Haoyu Wang, Xinliang Wang, Kele Xu, Hung Yeh, Doyeob Yeo, Yixuan Yuan, Yan Zeng , Xin Zhao, Julian Abbing, Jannes Adam, Nagesh Adluru, Niklas Agethen, Salman Ahmed, Yasmina Al Khalil, Mireia Alenyà, Esa Alhoniemi, Chengyang An, Talha Anwar, Tewodros Weldebirhan Arega, Netanell Avisdris, Dogu Baran Aydogan, Yingbin Bai, Maria Baldeon Calisto, Berke Doga Basaran, Marcel Beetz, Cheng Bian, Hao Bian, Kevin Blansit, Louise Bloch, Robert Bohnsack, Sara Bosticardo, Jack Breen, Mikael Brudfors, Raphael Brüngel, Mariano Cabezas, Alberto Cacciola, Zhiwei Chen, Yucong Chen, Daniel Tianming Chen, Minjeong Cho, Min-Kook Choi, Chuantao Xie Chuantao Xie, Dana Cobzas, Julien Cohen-Adad, Jorge Corral Acero, Sujit Kumar Das, Marcela de Oliveira, Hanqiu Deng, Guiming Dong, Lars Doorenbos, Cory Efird, Di Fan, Mehdi Fatan Serj, Alexandre Fenneteau, Lucas Fidon, Patryk Filipiak, René Finzel, Nuno R. Freitas, Christoph M. Friedrich, Mitchell Fulton, Finn Gaida, Francesco Galati, Christoforos Galazis, Chang Hee Gan, Zheyao Gao, Shengbo Gao, Matej Gazda, Beerend Gerats, Neil Getty, Adam Gibicar, Ryan Gifford, Sajan Gohil, Maria Grammatikopoulou, Daniel Grzech, Orhun Güley, Timo Günnemann, Chunxu Guo, Sylvain Guy, Heonjin Ha, Luyi Han, Il Song Han, Ali Hatamizadeh, Tian He, Jimin Heo, Sebastian Hitziger, SeulGi Hong, SeungBum Hong, Rian Huang, Ziyan Huang, Markus Huellebrand, Stephan Huschauer, Mustaffa Hussain, Tomoo Inubushi, Ece Isik Polat, Mojtaba Jafaritadi, SeongHun Jeong, Bailiang Jian, Yuanhong Jiang, Zhifan Jiang, Yueming Jin, Smriti Joshi, Abdolrahim Kadkhodamohammadi, Reda Abdellah Kamraoui, Inha Kang, Junghwa Kang, Davood Karimi, April Khademi, Muhammad Irfan Khan, Suleiman A. Khan, Rishab Khantwal, Kwang-Ju Kim, Timothy Kline, Satoshi Kondo, Elina Kontio, Adrian Krenzer, Artem Kroviakov, Hugo Kuijf, Satyadwyoom Kumar, Francesco La Rosa, Abhi Lad, Doohee Lee, Minho Lee, Chiara Lena, Hao Li, Ling Li, Xingyu Li, Fuyuan Liao, KuanLun Liao, Arlindo Limede Oliveira, Chaonan Lin, Shan Lin, Akis Linardos, Marius George Linguraru, Han Liu, Tao Liu, Di Liu, Yanling Liu, João Lourenço-Silva, Jingpei Lu, Jiangshan Lu, Imanol Luengo, Christina B. Lund, Huan Minh Luu, Yi Lv, Yi Lv, Uzay Macar, Leon Maechler, Sina Mansour L., Kenji Marshall, Moona Mazher, Richard McKinley, Alfonso Medela, Felix Meissen, Mingyuan Meng, Dylan Miller, Seyed Hossein Mirjahanmardi, Arnab Mishra, Samir Mitha, Hassan Mohy-ud-Din, Tony Chi Wing Mok, Gowtham Krishnan Murugesan, Enamundram Naga Karthik, Sahil Nalawade, Jakub Nalepa, Mohamed Naser, Ramin Nateghi, Hammad Naveed, Quang-Minh Nguyen, Cuong Nguyen Quoc, Brennan Nichyporuk, Bruno Oliveira, David Owen, Jimut Bahan Pal, Junwen Pan, Wentao Pan, Winnie Pang, Bogyu Park, Vivek Pawar, Kamlesh Pawar, Michael Peven, Lena Philipp, Tomasz Pieciak, Szymon Plotka, Marcel Plutat, Fattaneh Pourakpour, Domen Preložnik, Kumaradevan Punithakumar, Abdul Qayyum, Sandro Queirós, Arman Rahmim, Salar Razavi, Jintao Ren, Mina Rezaei, Jonathan Adam Rico, ZunHyan Rieu, Markus Rink, Johannes Roth, Yusely Ruiz-Gonzalez, Numan Saeed, Anindo Saha, Mostafa Salem, Ricardo Sanchez-Matilla, Kurt Schilling, Wei Shao, Zhiqiang Shen, Ruize Shi, Pengcheng Shi, Daniel Sobotka, Théodore Soulier, Bella Specktor Fadida, Danail Stoyanov, Timothy Sum Hon Mun, Xiaowu Sun, Rong Tao, Franz Thaler, Antoine Théberge, Felix Thielke, Helena Torres, Kareem A. Wahid, Jiacheng Wang, YiFei Wang, Wei Wang, Xiong Wang, Jianhui Wen, Ning Wen, Marek Wodzinski, Ye Wu, Fangfang Xia, Tianqi Xiang, Chen Xiaofei, Lizhan Xu, Tingting Xue, Yuxuan Yang, Lin Yang, Kai Yao, Huifeng Yao, Amirsaeed Yazdani, Michael Yip, Hwanseung Yoo, Fereshteh Yousefirizi, Shunkai Yu, Lei Yu, Jonathan Zamora, Ramy Ashraf Zeineldin, Dewen Zeng, Jianpeng Zhang, Bokai Zhang, Jiapeng Zhang, Fan Zhang, Huahong Zhang, Zhongchen Zhao, Zixuan Zhao, Jiachen Zhao, Can Zhao, Qingshuo Zheng, Yuheng Zhi, Ziqi Zhou, Baosheng Zou, Klaus Maier-Hein, Paul F. Jäger, Annette Kopp-Schneider, Lena Maier-Hein
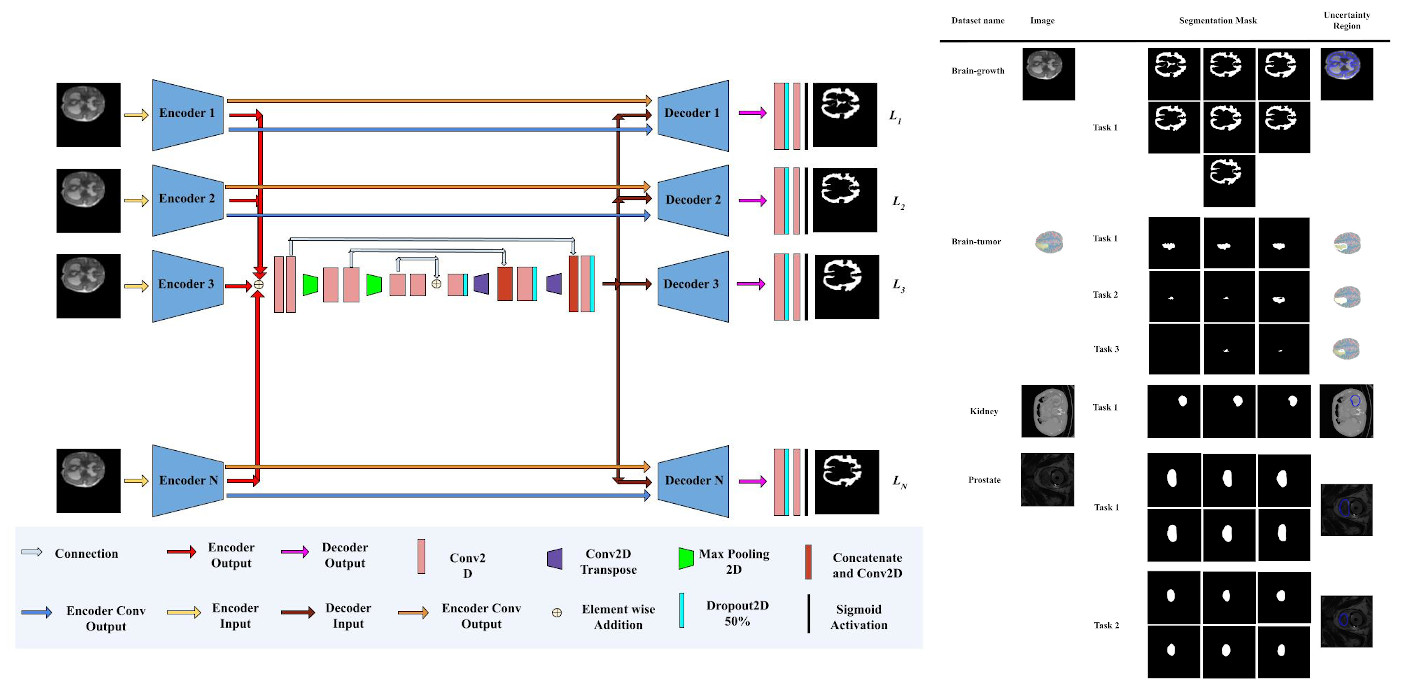
Holistic network for quantifying uncertainties in medical images
International MICCAI Brainlesion Workshop, BrainLes 2021, Brainlesion, Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. (2022)
Abstract: Variability in delineation is an inherent property for segmenting medical imagery, when images are annotated by a variety of expert annotators. Previous methods have used adversarial training, Monte-Carlo sampling, and dropouts, which might sometimes produce a wide range of segmentation masks that differ from the styles of mask produced by a set of expert annotators. State-of-the-art method uses multiple U-Nets to capture the individual delineations, but it is computationally demanding. To mitigate this problem, a holistic network containing N-Encoder and N-Decoder is proposed, which could individually model the variability of delineation produced by the expert annotators. This will help to create segmentation masks for different tasks of the same dataset through a single network by learning the common features of multiple Encoders via a common channel and passing those features to Decoder. These create one segmentation mask. All the masks are calculated by using weighted loss at each end of the Decoders that show excellent results for some datasets.
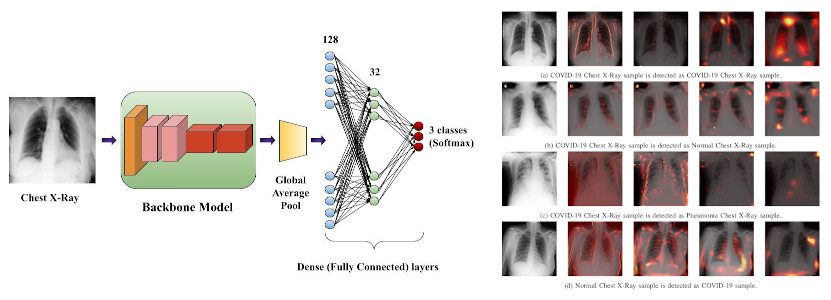
Classifying Chest X-Ray COVID-19 images via Transfer Learning
2021 Ethics and Explainability for Responsible Data Science (EE-RDS), published in IEEE Xplore. (2021)
Abstract: The internal behavior of Deep Neural Network architectures can be difficult to interpret. Certain architectures achieve impressive feats in a particular dataset while failing to show comparable performance in other datasets. Developing an architecture that performs well on a dataset can be a time-consuming affair and computationally intensive process. This study explains the effect of transfer learning by fine-tuning already available state-of-the-art architectures in different datasets and using them to classify Chest X-Ray images with high accuracy. Using transfer learning helps the model learn problem-specific features in a short period. It further shows that different models perform differently in a particular setting for a dataset. Ablation studies show that a combination of smaller structures that gives an overall better result may not give the best result in the combined model. In addition, the “belief” of the model for selecting a particular class is visualized in this study.
This material is presented to ensure timely dissemination of scholarly and technical work.
Copyright and all rights therein are retained by authors or by other copyright holders.
All persons copying this information are expected to adhere to the terms and constraints invoked by each author's copyright.
In most cases, these works may not be reposted without the explicit permission of the copyright holder.
 ORCID ID: 0000-0002-1206-7902
ORCID ID: 0000-0002-1206-7902